01. Project Overview
Quantifying Hippocampus Volume for Alzheimer's Progression
Background
Alzheimer's disease (AD) is a progressive neurodegenerative disorder that results in impaired neuronal (brain cell) function and eventually, cell death. AD is the most common cause of dementia. Clinically, it is characterized by memory loss, inability to learn new material, loss of language function, and other manifestations.
For patients exhibiting early symptoms, quantifying disease progression over time can help direct therapy and disease management.
A radiological study via MRI exam is currently one of the most advanced methods to quantify the disease. In particular, the measurement of hippocampal volume has proven useful to diagnose and track progression in several brain disorders, most notably in AD. Studies have shown a reduced volume of the hippocampus in patients with AD.
The hippocampus is a critical structure of the human brain (and the brain of other vertebrates) that plays important roles in the consolidation of information from short-term memory to long-term memory. In other words, the hippocampus is thought to be responsible for memory and learning (that's why we are all here, after all!)
 </small>](img/hippocampus-small.gif)
Hippocampus
Source: Life Science Databases (LSDB). Hippocampus. Images are from Anatomography maintained by Life Science Databases (LSDB). (2010). CC-BY-SA 2.1jp. Link
Humans have two hippocampi, one in each hemisphere of the brain. They are located in the medial temporal lobe of the brain. Fun fact - the word "hippocampus" is roughly translated from Greek as "horselike" because of the similarity to a seahorse observed by one of the first anatomists to illustrate the structure, but you can also see the comparison in the following image.
 </small>](img/hippocampus-and-seahorse-cropped.jpg)
Seahorse & Hippocampus
Source: Seress, Laszlo. Laszlo Seress' preparation of a human hippocampus alongside a sea horse. (1980). CC-BY-SA 1.0. Link
According to Nobis et al., 2019, the volume of hippocampus varies in a population, depending on various parameters, within certain boundaries, and it is possible to identify a "normal" range taking into account age, sex and brain hemisphere.
You can see this in the image below where the right hippocampal volume of women across ages 52 - 71 is shown.
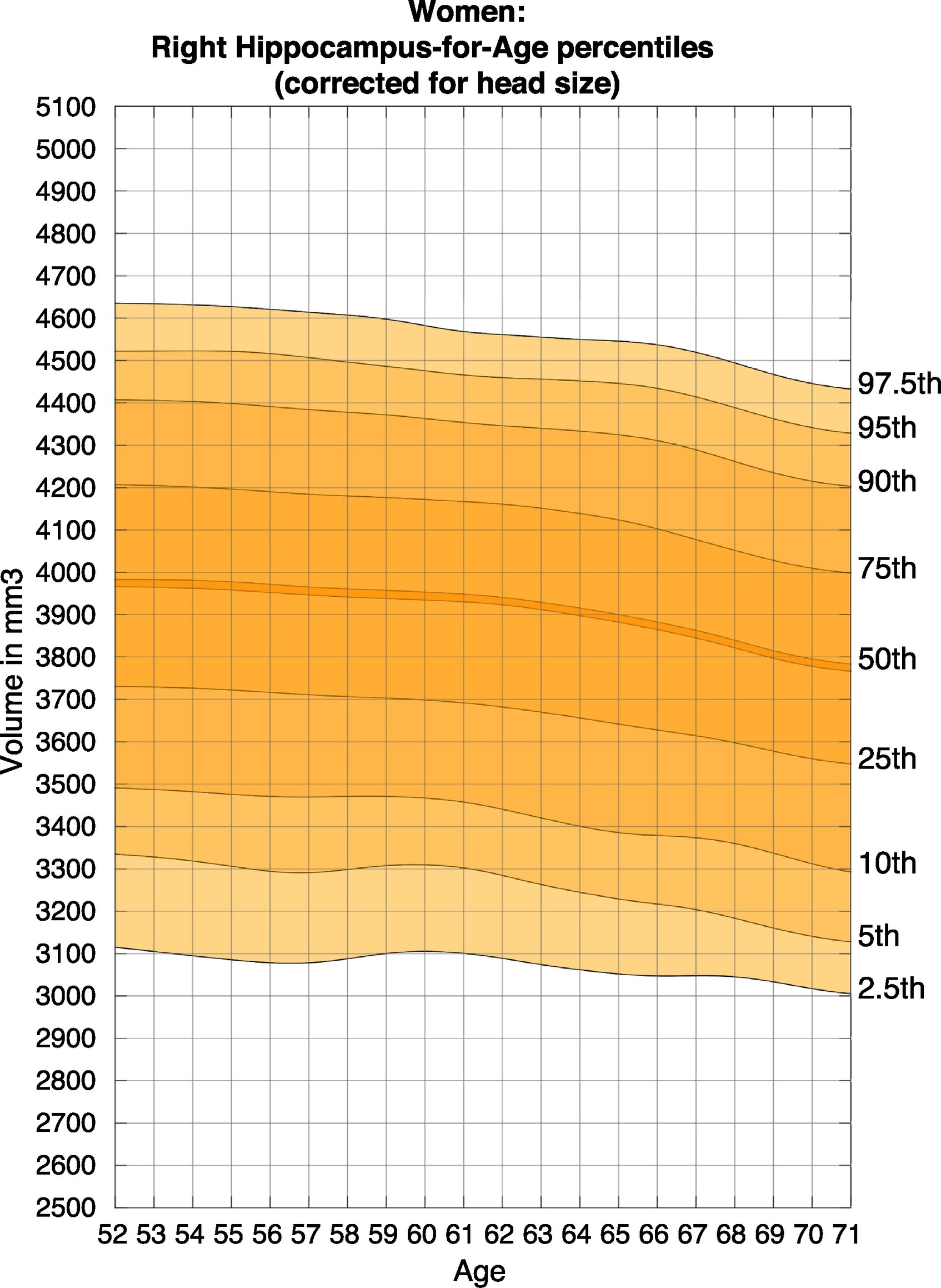
Nomogram - Female, Right Hippocampus Volume, Corrected for Head Size
Source: Nobis, L., Manohar, S.G., Smith, S.M., Alfaro-Almagro, F., Jenkinson, M., Mackay, C.E., Husain, M. Hippocampal volume across age: Nomograms derived from over 19,700 people in UK Biobank. Neuroimage: Clinical, 23(2019), pp. 2213-1582.
There is one problem with measuring the volume of the hippocampus using MRI scans, though - namely, the process tends to be quite tedious since every slice of the 3D volume needs to be analyzed, and the shape of the structure needs to be traced. The fact that the hippocampus has a non-uniform shape only makes it more challenging. Do you think you could spot the hippocampi in this axial slice below?
Axial slice of an MRI image of the brain
As you might have guessed by now, we are going to build a piece of AI software that could help clinicians perform this task faster and more consistently.
You have seen throughout the course that a large part of AI development effort is taken up by curating the dataset and proving clinical efficacy. In this project, we will focus on the technical aspects of building a segmentation model and integrating it into the clinician's workflow, leaving the dataset curation and model validation questions largely outside the scope of this project.
What You Will Build
In this project you will build an end-to-end AI system which features a machine learning algorithm that integrates into a clinical-grade viewer and automatically measures hippocampal volumes of new patients, as their studies are committed to the clinical imaging archive.
Fortunately you won't have to deal with full heads of patients. Our (fictional) radiology department runs a HippoCrop tool which cuts out a rectangular portion of a brain scan from every image series, making your job a bit easier, and our committed radiologists have collected and annotated a dataset of relevant volumes, and even converted them to NIFTI format!
You will use the dataset that contains the segmentations of the right hippocampus and you will use the U-Net architecture to build the segmentation model.
After that, you will proceed to integrate the model into a working clinical PACS such that it runs on every incoming study and produces a report with volume measurements.
The Dataset
We are using the "Hippocampus" dataset from the Medical Decathlon competition. This dataset is stored as a collection of NIFTI files, with one file per volume, and one file per corresponding segmentation mask. The original images here are T2 MRI scans of the full brain. As noted, in this dataset we are using cropped volumes where only the region around the hippocampus has been cut out. This makes the size of our dataset quite a bit smaller, our machine learning problem a bit simpler and allows us to have reasonable training times. You should not think of it as "toy" problem, though. Algorithms that crop rectangular regions of interest are quite common in medical imaging. Segmentation is still hard.
The Programming Environment
You will have two options for the environment to use throughout this project:
Udacity Workspaces
These are setup environments that contains all you need from the Local Environment section below that you can run directly on your web browser.
Local Environment
If you would like to run the project locally, you would need a Python 3.7+ environment with the following libraries for the first two sections of the project:
- PyTorch (preferably with CUDA)
- nibabel
- matplotlib
- numpy
- pydicom
- Pillow (should be installed with pytorch)
- tensorboard
In the 3rd section of the project we will be working with three software products for emulating the clinical network. You would need to install and configure:
- Orthanc server for PACS emulation
- OHIF zero-footprint web viewer for viewing images. Note that if you deploy OHIF from its github repository, at the moment of writing the repo includes a yarn script (
orthanc:up) where it downloads and runs the Orthanc server from a Docker container. If that works for you, you won't need to install Orthanc separately. - If you are using Orthanc (or other DICOMWeb server), you will need to configure OHIF to read data from your server. OHIF has instructions for this: https://docs.ohif.org/configuring/data-source.html
- In order to fully emulate the Udacity workspace, you will also need to configure Orthanc for auto-routing of studies to automatically direct them to your AI algorithm. For this you will need to take the script that you can find at
section3/src/deploy_scripts/route_dicoms.luaand install it to Orthanc as explained on this page: https://book.orthanc-server.com/users/lua.html - DCMTK tools for testing and emulating a modality. Note that if you are running a Linux distribution, you might be able to install dcmtk directly from the package manager (e.g.
apt-get install dcmtkin Ubuntu)
You can look at the rubric for this project here. Let's get started!